Announce the publication of a comprehensive Adjuvant Database to facilitate preclinical evaluation of vaccines and immunotherapeutics
Key Research Findings
- A transcriptome database was constructed for 25 types of vaccine adjuvants
- The characteristics of the adjuvants were reflected in the gene expression profiles
- Integration with toxicity database “Open TG-GATEs” enabled the prediction of adjuvanticity and toxicity
- The results of adjuvanticity and toxicity predictions using machine learning were validated by experiments
Abstract
Adjuvants are immune stimulants that enhance the efficacy of vaccines, but methods for evaluating their efficacy and safety are limited, making large-scale screening difficult. Therefore, we constructed the “Adjuvant Database (ADB)” containing comprehensive transcriptome data (*1) for 25 types of adjuvants across different species, organs, time points, and doses. This database was created using the same protocol as the widely used toxicity database “Open TG-GATEs (OTG),” enabling integrated analysis of both datasets. Each adjuvant exhibited a unique gene expression profile. By utilizing both databases and constructing a model to predict adjuvant activity and hepatotoxicity using machine learning (*2), we identified the adjuvant activity of colchicine in OTG and the hepatotoxicity of FK565, a synthetic immune stimulant derived from bacteria, in ADB through data analysis. This study demonstrates a new framework for adjuvant screening utilizing transcriptome information.
Details of Study
July 17, 2025 – National Institutes of Biomedical Innovation, Health and Nutrition (NIBN) and Department of Vaccine Science, The Institute of Medical Science, The University of Tokyo have developed the “Adjuvant Database (ADB),” which is expected to significantly contribute to the preclinical evaluation of the efficacy and safety of vaccine adjuvants and immunotherapy drugs. The findings of this study have been published in the August issue , which will be released on August 11, in the international scientific journal Cell Chemical Biology.
Adjuvants are essential components of vaccines that are necessary for vaccine antigen-specific immune responses. However, they are also being developed and researched as drugs that can enhance immune responses against infections, cancer, and allergies when used alone, making them a subject of growing interest. However, it is known that the components and mechanisms of action of adjuvants are diverse and complex, and there are no standardized methods for evaluating their efficacy and safety. As a result, it has been pointed out that the time required for market approval is longer than for other drugs, making drug discovery research challenging.
This time, an interdisciplinary research team led by, along with Dr. Ken Ishii, former Director of the Vaccine and Adjuvant Research Center, National Institutes of Biomedical Innovation, Health and Nutrition and currently a professor at the Department of Vaccine Science, The Institute of Medical Science, The University of Tokyo, and Dr. Kouji Kobiyama, Associate Professor at the same institute (at the time of the study; currently Associate Project Scientist, Department of Medicine, Vc-Health Sciences-Schools at the University of California, San Diego), along with Dr. Yayoi Natsume, Director of the AI Center for Health and Biomedical Research, and Dr. Jun Kunisawa, Deputy Director of the National Institute of Biomedical Innovation and Director of Microbial Research Center for Health and Medicine, National Institutes of Biomedical Innovation, Health and Nutrition have constructed an ADB by collecting comprehensive gene expression profiles for 25 major adjuvants using standardized methods across multiple animal species, organs, doses, and time points, and integrating them with the widely used toxicogenomics database OTG (Open TG-GATEs) (Figure 1).
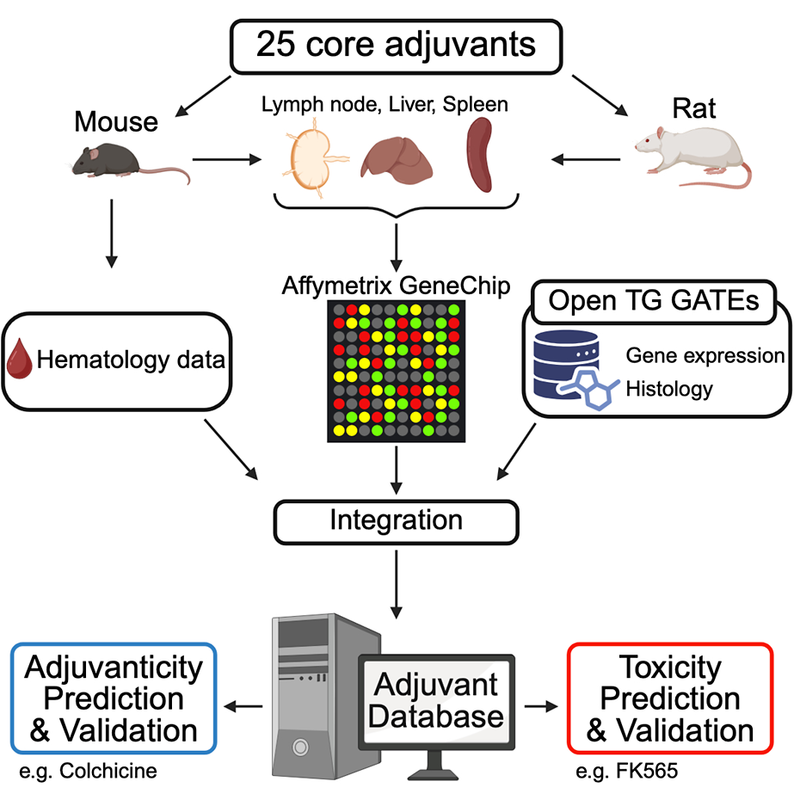
Figure 1 Establishment and Analysis of the Adjuvant Database (ADB)
These data can be easily viewed and analyzed using a user-friendly web application. This makes it possible to clearly show the gene expression patterns specific to each adjuvant, and the tendency for adjuvants with similar mechanisms of action to have similar gene expression profiles indicates that the characteristics of adjuvants are reflected in their gene expression patterns (Figure 2).
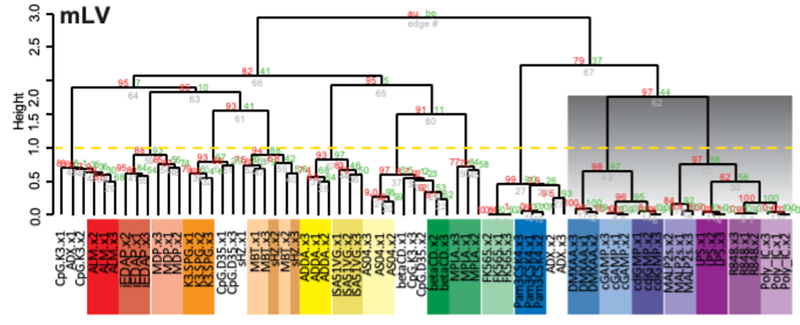
Figure 2 Adjuvant‑induced Gene Expression Profile in Mouse Liver
From this, it is thought that it is possible to infer the biological response induced by adjuvants from the ADB gene expression profile, and in fact, a group of genes correlated with blood cell counts was identified. The gene expression profiles of adjuvants collected in ADB contain information on their mechanisms of action and biological responses, and since data collection is performed using a unified protocol with OTG, it is possible to utilize these characteristics. As a specific example, a bidirectional prediction model for the efficacy and toxicity of candidate adjuvants and existing registered drugs as adjuvants was developed using machine learning.
Two novel insights were gained from this predictive model. The first was that, among the adjuvant candidate drugs in the ADB, FK565, a synthetic immune stimulant derived from bacteria, was predicted to exhibit liver toxicity based on the results of liver toxicity prediction using machine learning with OTG data. This was confirmed by experiments in mice and rats, which demonstrated strong toxicity inducing necrosis in the liver (Figure 3).
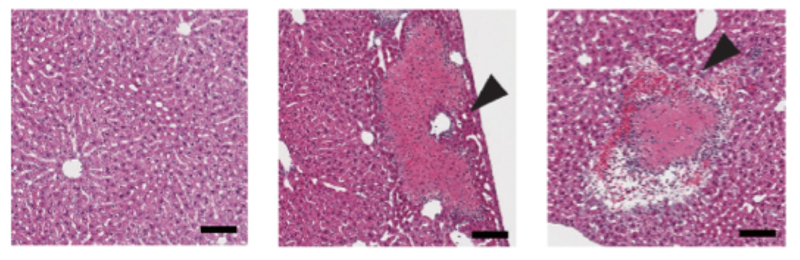
Figure 3 Necrosis in mouse liver following FK565 administration(from left: day 1, day 2, day 3 post‑treatment; scale bar = 100 µm)
Second, conversely, among the drugs registered in OTG, colchicine, which has long been used clinically as a preventive agent for gout attacks and a treatment for autoinflammatory diseases such as Mediterranean fever, was predicted by machine learning to be a potential adjuvant. In actual experiments using mice, colchicine was proven to function as an effective adjuvant for protein vaccines (Figure 4).
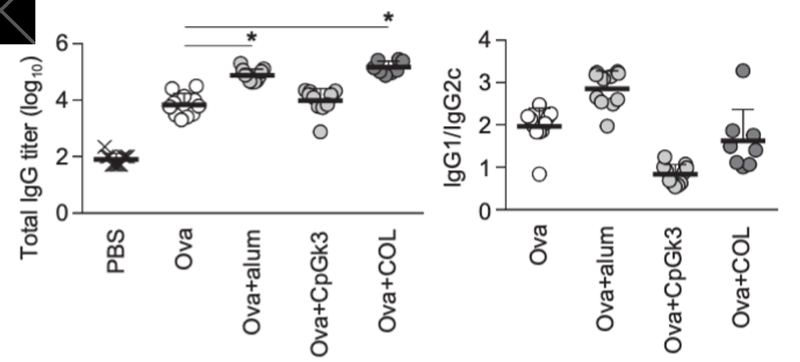
Figure 4 Colchicine‑induced enhancement of antibody production
The construction of ADB is expected to be widely used not only by academia but also by researchers around the world involved in vaccine and immunotherapy drug discovery and regulation, as it is a powerful data-driven screening system that accelerates the development of safer and more effective vaccines and can be utilized in parallel with actual experiments for important evaluations in non-clinical trials, such as toxicity.
For details of the research, please refer to the online edition of Cell Chemical Biology.
Article Information
Journal: Cell Chemical Biology
Title: An Adjuvant Database for preclinical evaluation of vaccines and immunotherapeutics
Authors: Yayoi Natsume-Kitatani, Kouji Kobiyama, Yoshinobu Igarashi, Taiki Aoshi, Noriyuki Nakatsu, Lokesh P. Tripathi, Junichi Ito, Johan Nyström-Persson, Yuji Kosugi, Rodolfo S. Allendes Osorio, Chioko Nagao, Burcu Temizoz, Etsushi Kuroda, Daron M. Standley, Hiroshi Kiyono, Kenji Nakanishi, Satoshi Uematsu, Isao Hamaguchi, Yasuhiro Yasutomi, Jun Kunisawa, Sho Yamasaki, Cevayir Coban, Hiroshi Yamada, Kenji Mizuguchi, and Ken J. Ishii* (*corresponding author)
Release date
Public release is scheduled for 11:00 AM Japan Standard Time on August 11, 2025.
Funding Acknowledgement
This work was supported by research grants from the Japan Agency for Medical Research and Development (AMED), under Research on Development of New Drugs, Division of Pharmaceutical Research and Development, covering Fiscal Years 2012–2016. The project was titled “Development of the next-generation adjuvants with innovative evaluation systems for safety and efficacy”, under grant numbers JP17ak0101068, JP18ak0101068, and JP17fm0208021. In addition, support was provided by the Japan Science and Technology Agency (JST) under Strategic Basic Research Programs (CREST), covering Fiscal Years 2018–2023, for the project titled “Immune response to, and regulation by, extracellular nucleic acids” (Grant number JPMJCR18H1).
Terminology
*1:Transcriptome data
Comprehensive measurements of which genes (DNA regions) are transcribed into messenger RNA in cells or tissues, and to what extent—used to profile gene expression on a genome-wide scale.
*2:Machine learning
A subfield of artificial intelligence (AI) that involves algorithms learning patterns or rules from data to perform predictive tasks or discover insights.
Contact for Inquiries
Planning Division, Department of Strategy and Research Support, NIBN
https://www.nibn.go.jp/en/contact/index.html
Press Release Material ![]() (559KB)
(559KB)![]()

トップページ「What's New」欄に表示する画像

Achievement / Events / News のいずれかを入力してください。
Achievement
