Laboratory of in silico design
Members
| Title | Name |
|---|---|
| Project Leader | Suyong Re |
| Deputy Project Leader | Yi-An Chen |
| Project Researcher | Kaushik Rahul |
| Postdoctoral Research Associate |
Jelang Muhammad Dirgantara, Mochammad Arfin |
| Trainee | Firoozeh Piroozmand, YOSHIYAMA Hayato, Chih-Yang Cheng |
| Administrative Assistant | TOJO Mitsuyo |
| Former member | NISHIMA Wataru |
Background and objectives
This project promotes a rational drug discovery approach grounded in the fundamental principles of life science. This is achieved by integrating informatics technology, which utilizes large-scale data analysis, with structural simulation technology, which elucidates the molecular recognition mechanisms between disease-related proteins and drug candidates from their molecular structure and dynamics.
Under close collaboration with internal teams, external research institutions, and universities, we aim to establish a drug design platform capable of addressing diverse modalities, including medium and large molecules, in addition to conventional small molecule compounds. The ultimate goal is the high-precision and rapid design of drug candidate molecules for any therapeutic targets.
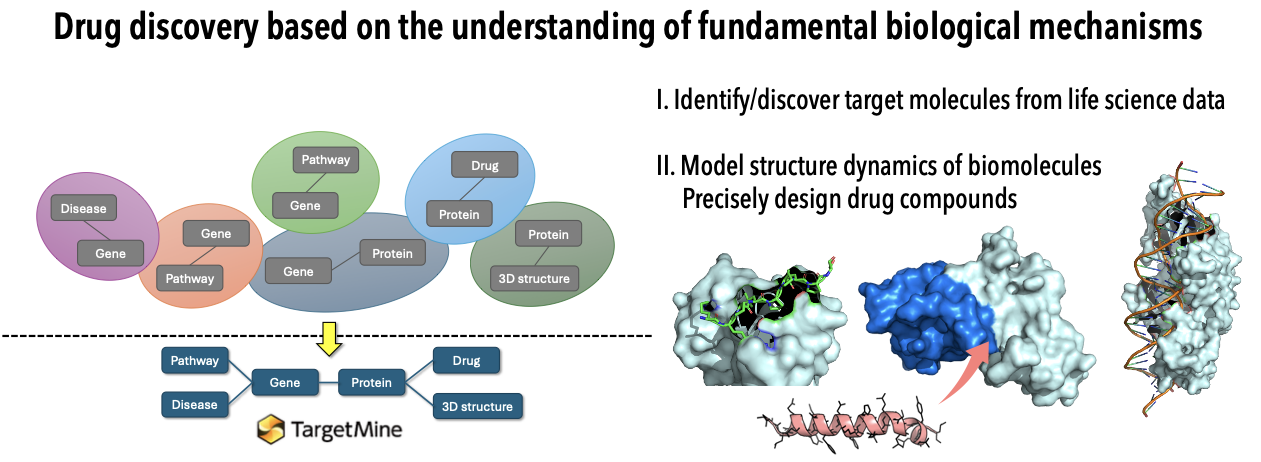
Overview of our research
Development of Data Integration Infrastructure and Application to Drug Discovery
We develop a platform that collects and analyzes publicly available life science data to conquer attractive drug targets currently unaddressed due to ineffective drug discovery strategies. By enabling detailed exploratory network analysis of disease-related target candidates, this platform aims to both accelerate drug design and significantly expand the drug discovery space.
Development of Structural Modeling Infrastructure and Application to Drug Discovery
We promote computational structure-based drug discovery (SBDD) technology by leveraging Artificial Intelligence (AI) and molecular simulation techniques to develop structural modeling technology that accurately visualizes the complex binding dynamics within biological systems, such as protein-protein interactions and other molecular recognition mechanisms between disease-related molecules. We aim to achieve a profound understanding of biomolecular recognition and, based on that knowledge, realize rational and innovative drug design.
References:
- Yi-An Chen, Hitoshi Kawashima, Jonguk Park, et al. “NIBN Japan Microbiome Database, a Database for Exploring the Correlations between Human Microbiome and Health.” Scientific Reports 15, (2025): 19640. https://doi.org/10.1038/s41598-025-04339-z
- Yi-An Chen, Rodolfo S Allendes Osorio, and Kenji Mizuguchi. “TargetMine 2022: A New Vision into Drug Target Analysis.” Bioinformatics 38, (2022): 4454–56. https://doi.org/10.1093/bioinformatics/btac507
- Yuhi Hosoe, Yohei Miyanoiri, Suyong Re, et al. “Structural Dynamics of the N‐terminal SH2 Domain of PI3K in Its Free and CD28 ‐bound States.” The FEBS Journal, 290, (2022): 2366-2378. https://doi.org/10.1111/febs.16666
- Daiki Yamane, Satsuki Onitsuka, Suyong Re, et al. “Selective Covalent Targeting of SARS-CoV-2 Main Protease by Enantiopure Chlorofluoroacetamide.” Chemical Science 13, (2022): 3027–34. https://doi.org/10.1039/D1SC06596C
- Taro Tsuji, Kayoko Hashiguchi, Mana Yoshida, et al. “α-Amino Acid and Peptide Synthesis Using Catalytic Cross-Dehydrogenative Coupling.” Nature Synthesis 1, (2022): 304–12. https://doi.org/10.1038/s44160-022-00037-0
- Suyong Re, and Kenji Mizuguchi. “Glycan Cluster Shielding and Antibody Epitopes on Lassa Virus Envelop Protein.” The Journal of Physical Chemistry B 125, (2021): 2089–97. https://doi.org/10.1021/acs.jpcb.0c11516
Contact information
| suyongre[at]nibn.go.jp (Replace [at] by @) |
Links
Artificial Intelligence Center for Health and Biomedical Research
NIBN JMD (Japan Microbiome Database)
GENESIS (GENeralized-Ensemble SImulation System)
- 2025
- 2024

トップページ「What's New」欄に表示する画像

Achievement / Events / News のいずれかを入力してください。
News
